Managing Datasets
3D image datasets are at the heart of webKnossos. Import datasets by uploading them directly via the UI or by using the file system (self-hosted instances only). Configure the dataset defaults and permissions to your specification. Share your datasets with the public or with selected users.
Importing Datasets
Uploading through the web browser
To import a dataset, you can use the upload functionality within webKnossos. For that, click "Add Dataset" in the dashboard. Then, drag your data into the form and ensure all form fields are filled.
webKnossos uses the WKW-format internally to display your data. If your data is already in WKW you can simply drag your folder (or zip archive of that folder) into the upload view. If your data is not in WKW, you can either: - upload the data to webknossos.org where it will be automatically converted to WKW - or convert your data manually to WKW.
Read more about the Data Formats we support and how they should be structured when uploading them.
Once the data is uploaded (and potentially converted) you can to go the Dataset Settings to doublecheck important properties (such as the dataset scale) or to make it public.
Convert Datasets
If your dataset is not yet in WKW format and you don't want to use webknossos.org for the conversion, you can also convert it manually. The webKnossos Cuber is a tool that can convert many formats to WKW. Read more in the tools section of the data formats documentation.
Uploading through the File System (Self-Hosted Instances Only)
On self-hosted instances, large datasets can be efficiently imported by placing them directly in the file system:
- Place the dataset at
<webKnossos directory>/binaryData/<Organization name>/<Dataset name>. For example/opt/webknossos/binaryData/Springfield_University/great_dataset. - Go to the dataset view on the dashboard
- Use the refresh button on the dashboard or wait for webKnossos to detect the dataset (up to 10min)
- Click
Importfor your new dataset - Complete the Import screen
Using Symbolic Links (Self-Hosted Instances Only)
You can also use symbolic links to import your data into webKnossos. However, when using Docker, the targets of the link also need to be available to the container through mounts.
For example, you could have a link from /opt/webknossos/binaryData/sample_organization/awesome_dataset to /cluster/path/to/dataset123.
In order to make this dataset available to the Docker container, you need to add /cluster as another volume mount.
You can add this directly to the docker-compose.yml:
...
services:
webknossos:
...
volumes:
- ./data:/srv/webknossos/binaryData
- /cluster:/cluster
...
Importing in webKnossos
The Import screen allows you to set some properties of your datasets. Many properties such as available layers, bounding boxes and datatypes can be detected automatically. Some properties may require your manual input, though. Most of the time these are scale which represents the physical size of one voxel in nanometers and largestSegmentId of a segmentation layer.
Once you entered the required properties, you can click the Import button to complete the process.
The dataset is now ready to use.
Info
If you uploaded the dataset along with a datasource-properties.json metadata file the dataset will be imported automatically without any additional manual steps.
Sample Datasets
A list of sample datasets is provided with webKnossos. Click Add a Sample Dataset on the upload page to access it and choose datasets to be added and imported automatically. The three sample datasets currently available are:
-
Sample_e2006_wkw: https://static.webknossos.org/data/e2006_wkw.zip Raw SBEM data and segmentation (sample cutout, 120MB). Connectomic reconstruction of the inner plexiform layer in the mouse retina. M Helmstaedter, KL Briggman, S Turaga, V Jain, HS Seung, W Denk. Nature. 08 August 2013. https://doi.org/10.1038/nature12346
-
Sample_FD0144_wkw: https://static.webknossos.org/data/FD0144_wkw.zip Raw SBEM data and segmentation (sample cutout, 316 MB). FluoEM, virtual labeling of axons in three-dimensional electron microscopy data for long-range connectomics. F Drawitsch, A Karimi, KM Boergens, M Helmstaedter. eLife. 14 August 2018. https://doi.org/10.7554/eLife.38976
-
Sample_MPRAGE_250um: https://static.webknossos.org/data/MPRAGE_250um.zip MRI data (250 MB). T1-weighted in vivo human whole brain MRI dataset with an ultrahigh isotropic resolution of 250 μm. F Lüsebrink, A Sciarra, H Mattern, R Yakupov, O Speck. Scientific Data. 14 March 2017. https://doi.org/10.1038/sdata.2017.32
Edit Dataset
You can edit the properties of a dataset at any time. In addition to the required properties that you need to fill in during import, there are more advanced properties that you can set. This screen is similar to the Import screen and split into three tabs:
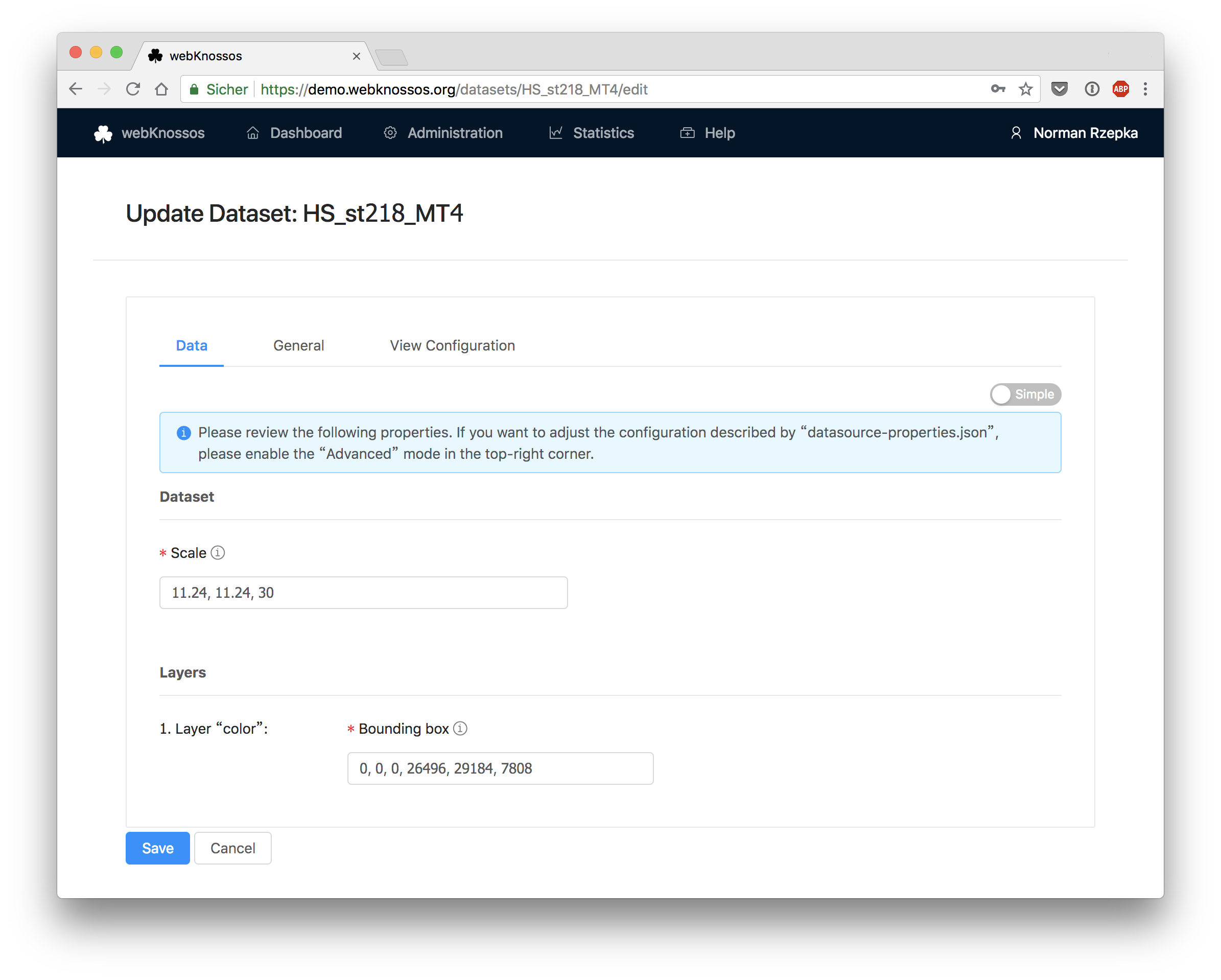
Data
Scale: The physical size of a voxel in nanometers, e.g.11, 11, 24Bounding Box: The position and extents of the dataset layer in voxel coordinates. The format isx,y,z,x_size,y_size,z_sizeor respectivelymin_x,min_y,min_z,(max_x-min_x),(max_y-min_y),(max_z-min_z).Largest Segment ID: The highest ID that is currently used in the respective segmentation layer. This is required for volume annotations where new objects with incrementing IDs are created. Only applies to segmentation layers.
The Advanced view lets you edit the underlying JSON configuration directly.
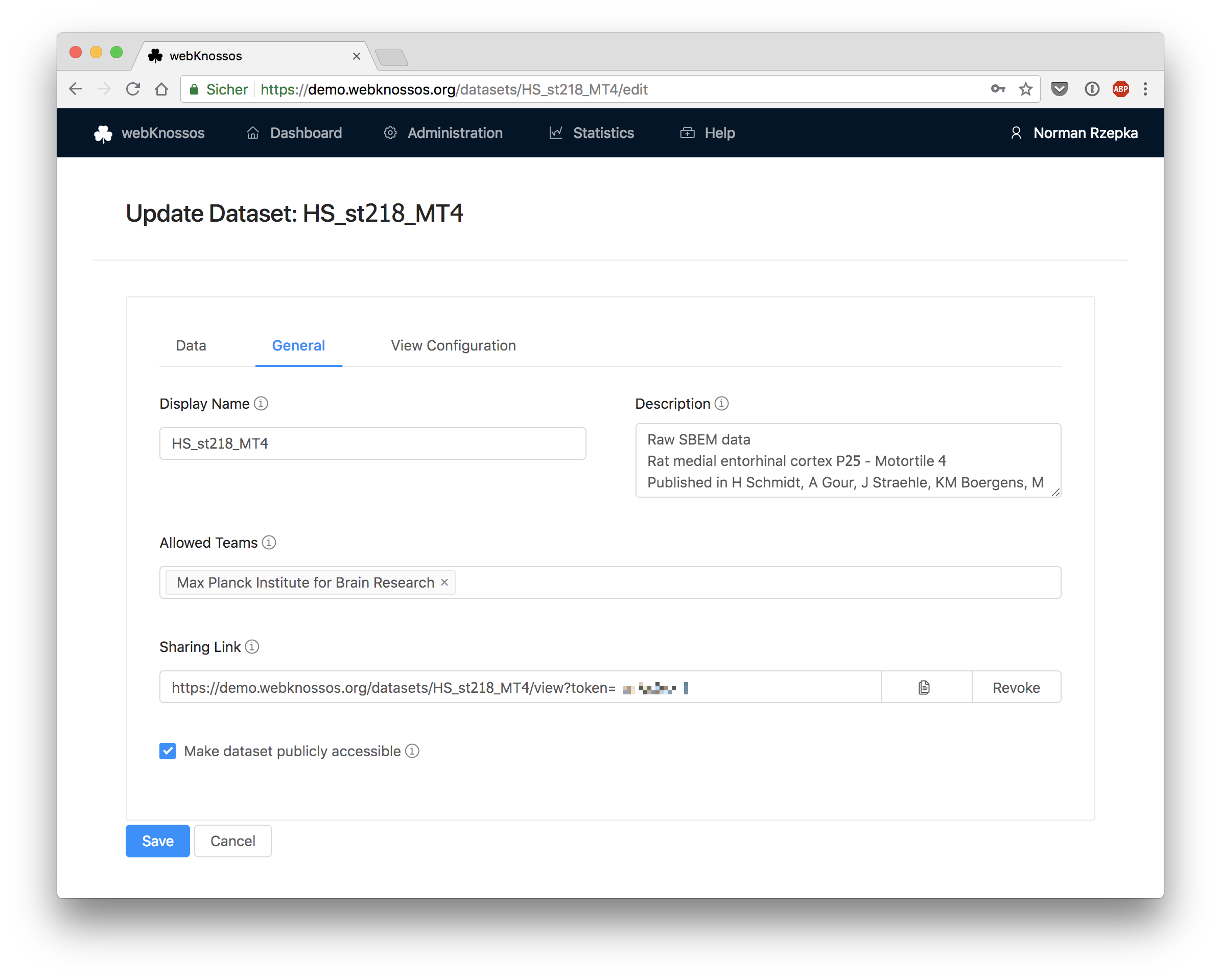
General
Display Name: Used as the name of the dataset in the Gallery view.Description: Contains more information about your datasets including authors, paper reference, descriptions. Supports Markdown formatting. The description will be featured in the Gallery view as well.Teams allowed to access this dataset: Defines which teams of your organization have access to this dataset. By default no team has access but admins and team managers can see and edit the dataset.Visibility: Lets you make the dataset available to the general public and shows it in the public Gallery view. This will enable any visitor to your webKnossos instance to view the data, even unregistered users.Sharing Link: A special URL which allows any user to view your dataset that uses this link. Because of the included random token, the link cannot be guessed by random visitors. You may also revoke the random token and create a new one when you don't want previous users to access your data anymore. Read more in the Sharing guide.
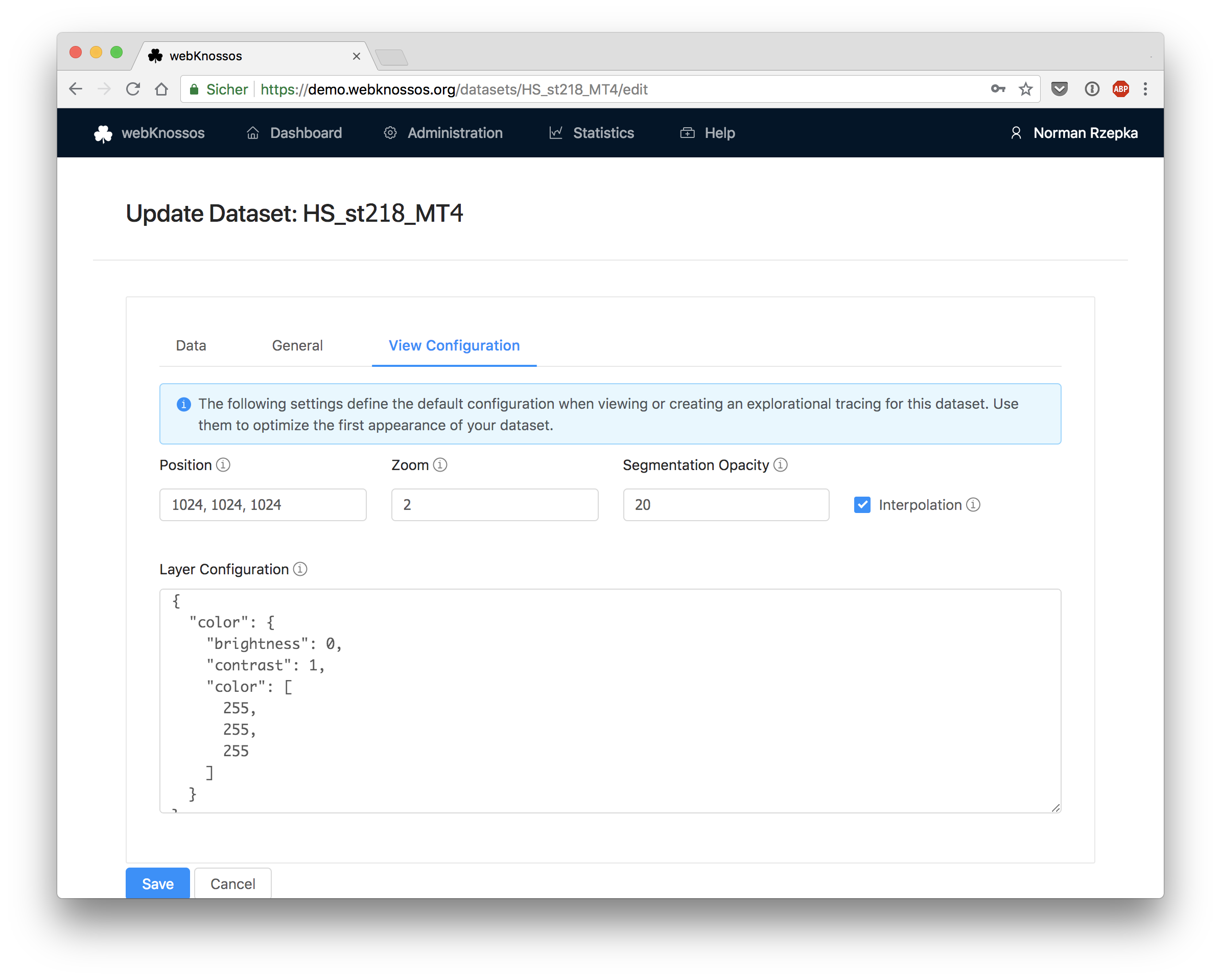
View Configuration
Position: Default position of the dataset in voxel coordinates. When opening the dataset, users will be located at this position.Zoom: Default zoom.Segmentation Pattern Opacity: Default opacity of the patterns rendered inside segmentation cells.Interpolation: Whether interpolation should be enabled by default.Layer Configuration: This is an advanced feature to control the default settings (e.g. alpha, color, intensity range) per layer. It needs to be configured in a JSON format.
View Configuration Hierarchy
There are two ways to set default View Configuration Settings:
- inside the datasource_properties.json
- in the Edit View for Datasets
The View Configuration from the Edit View takes precedence over the datasource_properties.json.
You don't have to set complete View Configurations in neither option, as webKnossos will fill missing attributes with sensible defaults.
These View Configurations impact the first appearance of the Dataset for all users.
Each user can further customize their View Configuration in the Annotation UI Settings
Dataset Sharing
Read more in the Sharing guide
Using External Datastores
The system architecture of webKnossos allows for versatile deployment options where you can install a dedicated datastore server directly on your lab's cluster infrastructure. This may be useful when dealing with large datasets that should remain in your data center. Please contact us or write a post, if you require any assistance with your setup.
scalable minds also offers a dataset alignment tool called Voxelytics Align. Learn more.